Featured Projects
HMDB molecular network amalgamation
Building of a graph database with Neo4J and MongoDB using Java, R, and D3.js. Metabolic pathway relationships were built using SMPDB with visualizations provided on the basis of chemical classifications. The tool is designed to extract sub-networks determined by expression data.
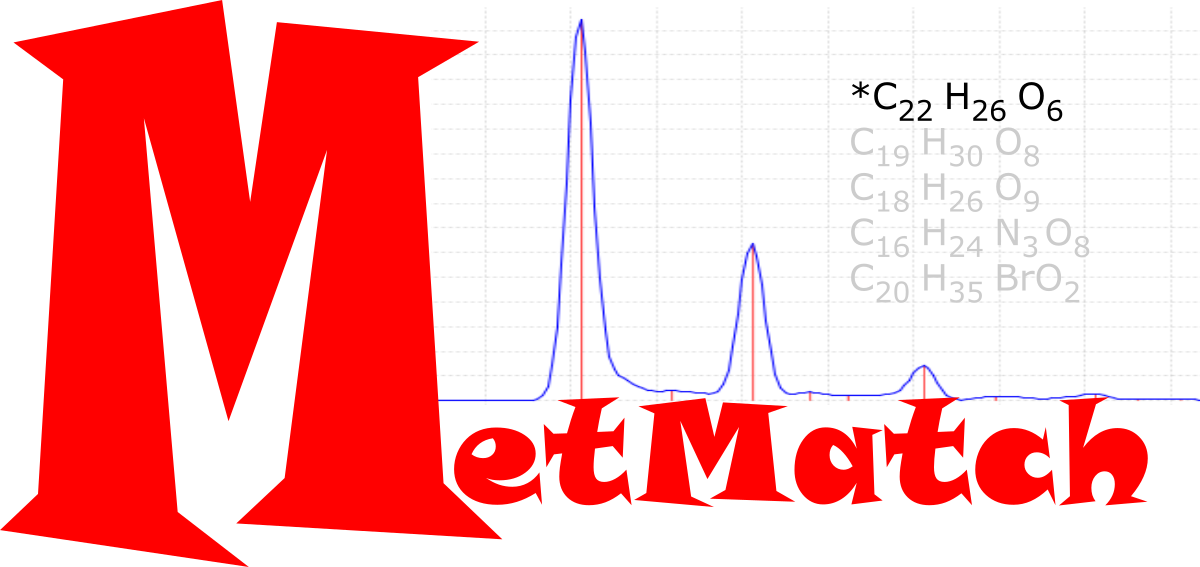
Rapid Lewis formula identification from static databases using isotopic distributions
Using theoretical isotope distrubutions of chemical formulae, we assembled a self correcting search algorithm via a global model method to improve accuracy and speed of molecular formula assignment.
Check it out